GROMACS-SWAXS
SAXS/WAXS/SANS calculations using all-atom MD simulations and SAXS-driven MD simulations with a modified GROMACS software
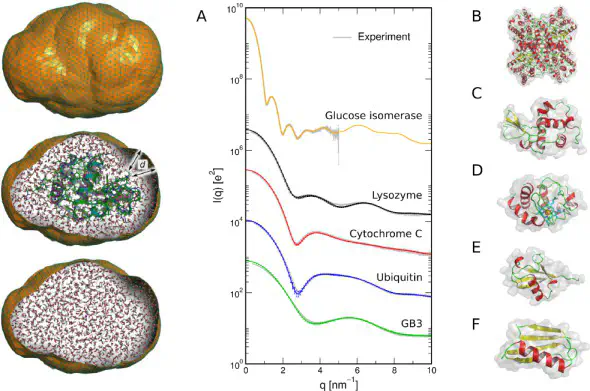
Our modification of GROMACS for computing small- and wide-angle X-ray or neutron scattering curves (SAXS/SANS), small-angle neutron scattering curves (SANS), and for doing SAXS/SANS-driven molecular dynamics simulations.
We provide a modified source code of GROMACS for the analysis of small- and wide-angle X-ray scattering data (SWAXS) as well as small-angle neutron scattering data (SANS). The code may be used to
- Compute SWAXS/SANS curves using all-atom explicit-solvent MD simulations, and
- to carry out SWAXS/SANS-driven simulations, i.e., to carry out structure refinement against SWAXS/SANS data using MD simulations.
Source code
The current source code is a modification of GROMACS 2021.7 and is available at GitLab. For compiling the code, please follow GROMACS-SWAXS documentation. Please note that this code is not official GROMACS, hence the GROMACS developers are not responsible for this extension. License: The code is distributed under the GNU Lesser General Public License (LGPL).
Web server
For computing only a few SWAXS curves our web server WAXSiS may be more suitable. WAXSiS runs a short explicit-solvent MD simulation from an uploaded PDB structure and subsequently computes the SWAXS curve. WAXSiS also allows the upload of a trajectory file. For more extensive calculations, however, the webserver gets cumbersome, hence you may better download the code and run the calculations on your computer.
Documentation and tutorials
The documentation can be found here. If you run into problems, please do not hesitate to contact us. We want you to use our software, so we’re happy to help.
Relevant publications
Overview articles
SWAXS/WAXS curve predictions from MD simulations
SWAXS-driven MD simulations
(structure refinement against SWAXS curves)