GROMACS GAFF topologies for 26 steroid compounds
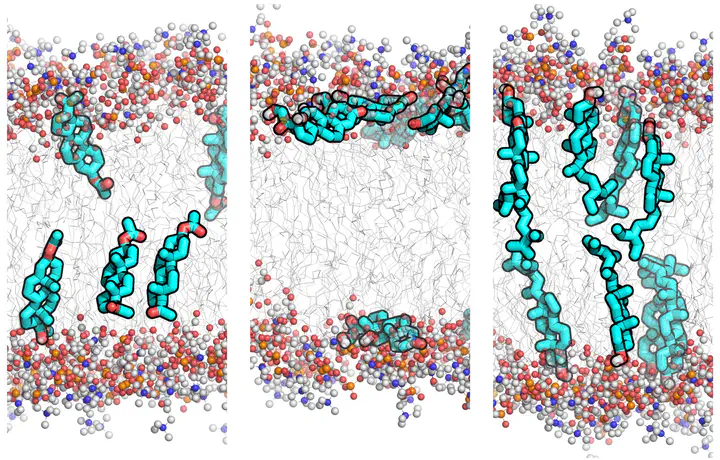
Simulation parameters (topologies) for the General Amber Force Field (GAFF) of 26 typical steroid compounds were derived and refined against experimental membrane/water partition coefficients.
On this site, we provide force field topology files in GROMACS format for 26 steroid compounds.
The atom types (Lennard-Jones parameters) are standard GAFF atoms. The partial charges were optimized to reproduce partition coefficients between a POPC membrane (modelled with Slipids) and TIP3P water. The topologies were not validated against any other data, such as protein binding affinities, so use them with caution.
Methods summary: The topologies were created with AmberTools14 using GAFF atom types and RESP charges. The electrostatic potential used for the RESP fit was calculated with Gaussian using HF/6-31G* (HF/6-31+G* for the anionic compounds) level of theory. The charges were optimized to reproduce experimentally measured partition coefficients between a POPC membrane and water ($K_{POPC}$), as follows. Two sets of RESP charges were derived from Hartree-Fock calculations: either in vacuum ($Q_{vac}$), or in a continuum water model ($Q_{wat}$). Then, for each steroid, the partial charges were linearly interpolated between the two sets, to best match the $K_{POPC}$ values, i.e.,
$Q_{opt} = w_{vac} Q_{vac} + (1-w_{vac})Q_{wat}$
where $w_{vac}$ was restricted to the interval [0, 1]. For steroids, for which KPOPC could not be obtained (due to solubility problems), the parameter $w_{vac}$ was guessed from steroids with similar substitutions. All $w_{vac}$ values are listed in this file. The topolgies were converted to Gromacs format by ACPYPE. For more details please refer to the reference below.
These files come without any warranty of any kind, to the extent applicable by local law.
If you use these files for publication, please cite
Please note: We discourage the use of the cholesterol GAFF topology, since many lipid force fields provide optimized cholesterol model. The model is here only provided for the sake of completeness.
| Steroid | Fromula | Structure file | Topology file |
|---|---|---|---|
| Aldosterone |  | ALD.pdb | ALD.itp |
| 4-Androstenedione |  | ASD.pdb | ASD.itp |
| β-Estradiol |  | BED.pdb | BED.itp |
| β-Sitosterol |  | BSL.pdb | BSL.itp |
| Cholesterol (discouraged) |  | chol_gaff.pdb | chol_gaff.itp |
| Hydrocortisone (Cortisol) |  | CTL.pdb | CTL.itp |
| Cortisone |  | CTN.pdb | CTN.itp |
| Corticosterone (17-deoxycortisol) |  | CTS.pdb | CTS.itp |
| Dehydroergosterol |  | DEL.pdb | DEL.itp |
| Dehydroepiandrosterone |  | DHEA.pdb | DHEA.itp |
| Dehydroepiandrosterone sulfate |  | DHEAS.pdb | DHEAS.itp |
| Dexamethasone |  | DMN.pdb | DMN.itp |
| Dihydrotestosterone |  | DTSN.pdb | DTSN.itp |
| Ethinylestradiol |  | EEL.pdb | EEL.itp |
| Estrone |  | ESN.pdb | ESN.itp |
| Estriol |  | ETL.pdb | ETL.itp |
| Fludrocortisone acetate |  | FCA.pdb | FCA.itp |
| Levonorgestrel |  | LGL.pdb | LGL.itp |
| Methandrostenolone |  | MAL.pdb | MAL.itp |
| Pregnenolone |  | PGN.pdb | PGN.itp |
| Pregnenolone acetate |  | PGNA.pdb | PGNA.itp |
| Pregnenolone sulfate |  | PGNS.pdb | PGNS.itp |
| Progesterone |  | PGS.pdb | PGS.itp |
| Prednisone |  | PNN.pdb | PNN.itp |
| Tetrahydrodeoxycorticosterone |  | TDOC.pdb | TDOC.itp |
| Testosterone |  | TSN.pdb | TSN.itp |
All topologies are also available with non-optimized charges, i.e. with the original HF/6-31G* (HF/6-31+G* for the anionic compounds) RESP charges obtained in vacuum. download