DNA opening during transcription initiation by RNA polymerase II in atomic detail
Abstract
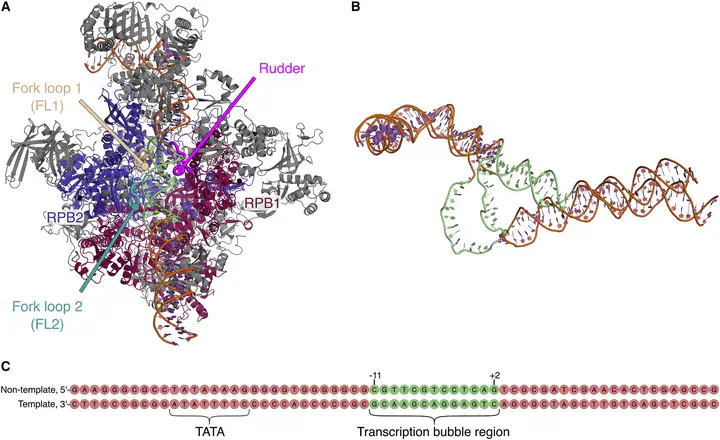
RNA polymerase II (RNAP II) synthesizes RNA by reading the DNA code. During transcription initiation, RNAP II opens the double-stranded DNA to expose the DNA template to the active site. The molecular interactions driving and controlling DNA opening are not well understood. We used all-atom steered molecular dynamics simulations to derive a continuous pathway of DNA opening in human RNAP II, involving a 55 Å DNA strand displacement and a nearly 360° DNA helix rotation. To drive such large-scale transitions, we used a combination of RMSD-based collective variables, a newly designed rotational coordinate, and a path collective variable. The simulations reveal extensive interactions of the DNA with three conserved protein loops near the active site, namely with the rudder, fork loop 1, and fork loop 2. According to the simulations, DNA–protein interactions support DNA opening by a twofold mechanism; they catalyze DNA opening by attacking Watson-Crick hydrogen bonds, and they stabilize the open DNA bubble by the formation of a wide set of DNA–protein salt bridges.