Small-Angle X-ray Scattering Curves of Detergent Micelles: Effects of Asymmetry, Shape Fluctuations, Disorder, and Atomic Details
Abstract
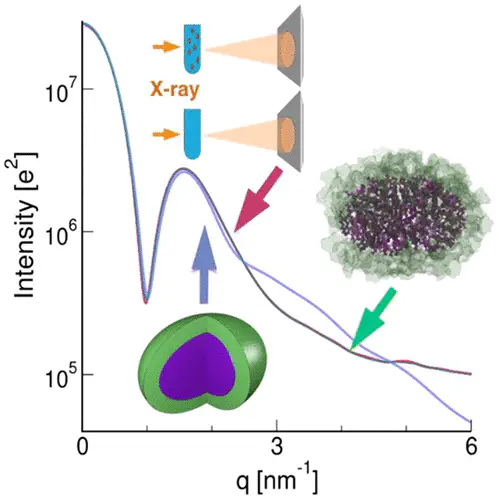
Small-angle X-ray scattering (SAXS) is a widely used experimental technique, providing structural and dynamic insight into soft-matter complexes and biomolecules under near-native conditions. However, interpreting the one-dimensional scattering profiles in terms of three-dimensional structures and ensembles remains challenging, partly because it is poorly understood how structural information is encoded along the measured scattering angle. We combined all-atom SAXS-restrained ensemble simulations, simplified continuum models, and SAXS experiments of a n-dodecyl-β-d-maltoside (DDM) micelle to decipher the effects of model asymmetry, shape fluctuations, atomic disorder, and atomic details on SAXS curves. Upon interpreting the small-angle regime, we find remarkable agreement between (i) a two-component triaxial ellipsoid model fitted against the data and (ii) a SAXS-refined all-atom ensemble. However, continuum models fail at wider angles, even if they account for shape fluctuations, disorder, and asymmetry of the micelle. We conclude that modeling atomic details is mandatory for explaining SAXS curves at wider angles.