Free-Energy Calculations of Pore Formation in Lipid Membranes
Abstract
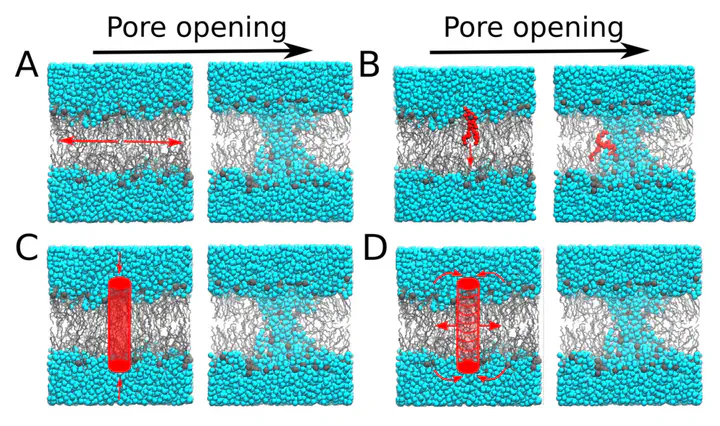
Aqueous pores over lipid membranes are biologically significant transient structures, and play important roles in membrane permeation, membrane fusion, antimicrobial peptide activity, and controlled transport of drug molecules and ions across cellular boundaries. Over the last 20 years, atomic and coarse-grained simulations have been used extensively to model the formation of transmembrane pores, and have hence provided detailed insight into the structures of open pores and into pathways of pore formation. Various perturbations were imposed in silico to drive pore formation, including electric fields, membrane tension, and membrane active peptides. Accurate free-energy calculations of pore formation, which can provide quantitative understanding of transmembrane pores, have remained challenging, in part due to the lack of good reaction coordinates (RCs). In this chapter, we review methods for free-energy calculations of pore formation, with a focus on RCs that have been proposed to calculate free-energy profiles for pore formation from molecular dynamics simulations.
In Biomembrane Simulations: Computational Studies of Biological Membranes, edited by Max Berkowitz.
Series in Computational Biophysics, CRC Press Taylor and Francis