Simulations of Pore Formation in Lipid Membranes: Reaction Coordinates, Convergence, Hysteresis, and Finite-Size Effects
Abstract
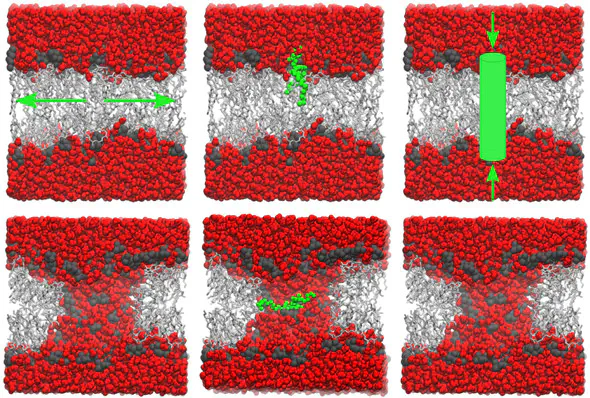
Transmembrane pores play an important role in various biophysical processes such as membrane permeation, membrane fusion, and antimicrobial peptide activity. In principal, all-atom molecular dynamics (MD) simulations provide an accurate model of pore formation in lipid membranes. However, the free energy landscape of transmembrane pore formation remains poorly understood, partly because potential of mean force (PMF) calculations of pore formation strongly depend on the choice of the reaction coordinate. In this study, we used umbrella sampling to compute PMFs for pore formation using three different reaction coordinates, namely, (i) a coordinate that steers the lipids in the lateral direction away from the pore center, (ii) the distance of a single lipid phosphate group from the membrane center, and (iii) the average water density inside a membrane-spanning cylinder. Our results show that while the three reaction coordinates efficiently form pores in membranes, they suffer from strong hysteresis between pore-opening and pore-closing simulations, suggesting that they do not restrain the systems close to the transition state for pore formation. The two reaction coordinates that act via restraining the lipids lead to more pronounced hysteresis compared with the coordinate acting on the water molecules. By comparing PMFs computed from membranes with different numbers of lipids, we observed significant artifacts from the periodic boundary conditions in small simulation systems. Further analysis suggests that the formation and disruption of a continuous hydrogen-bonding network across the membrane corresponds to the transition state for pore formation. Our study provides molecular insights into the critical steps of transmembrane pore formation, and it may guide the development of efficient reaction coordinates for pore formation.