WAXSiS: a web server for the calculation of SAXS/WAXS curves based on explicit-solvent molecular dynamics
Abstract
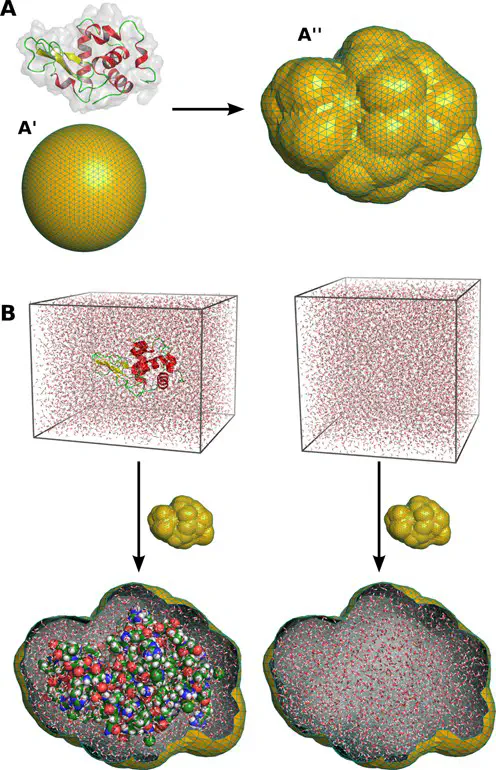
Small- and wide-angle X-ray scattering (SWAXS) has evolved into a powerful tool to study biological macromolecules in solution. The interpretation of SWAXS curves requires their accurate predictions from structural models. Such predictions are complicated by scattering contributions from the hydration layer and by effects from thermal fluctuations. Here, we describe the new web server WAXSiS (WAXS in solvent) that computes SWAXS curves based on explicit-solvent all-atom molecular dynamics (MD) simulations (http://waxsis.uni-goettingen.de http://waxsis.uni-saarland.de). The MD simulations provide a realistic model for both the hydration layer and the excluded solvent, thereby avoiding any solvent-related fitting parameters, while naturally accounting for thermal fluctuations.