BindFlow: A Free, User-Friendly Pipeline for Absolute Binding Free Energy Calculations Using Free Energy Perturbation or MM(PB/GB)SA
Abstract
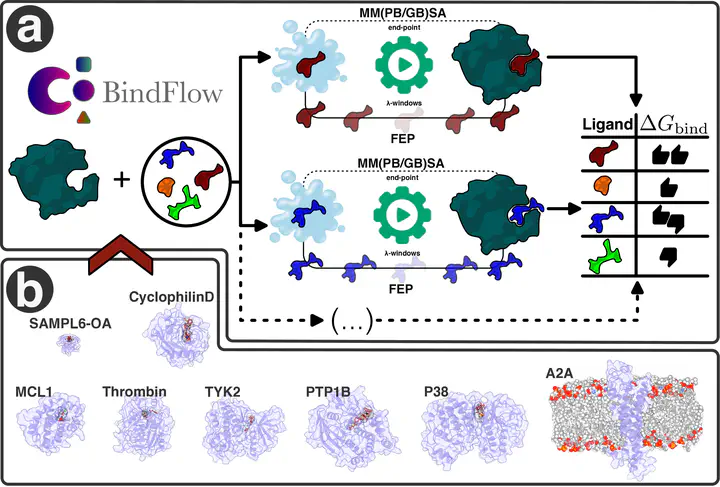
We present BindFlow, a Python-based software for automated absolute binding free energy (ABFE) calculations at the free energy perturbation (FEP) or at the molecular mechanics Poisson–Boltzmann/generalized Born surface area [MM(PB/GB)SA] level of theory. BindFlow is free, open-source, user-friendly, and easily customizable, runs on workstations or distributed computing platforms, and provides extensive documentation and tutorials. BindFlow uses GROMACS as a molecular dynamics engine and provides built-in support for the small-molecule force fields GAFF, OpenFF, and Espaloma, as well as support for user-provided custom force fields. We test BindFlow by computing affinities for 139 receptor–ligand pairs, involving eight different targets, including six soluble proteins, one membrane protein, and one nonprotein host–guest system. We find that the agreement of BindFlow predictions with experiments is overall similar to gold standards in the field. Interestingly, we find that MM(PB/GB)SA achieves correlations that, for some systems and force fields, approach those obtained with FEP while requiring only a fraction of the computational cost. This study establishes BindFlow as a validated and accessible tool for ABFE calculations.