Computational Biophysics Group
Welcome to the Computational Biophysics Group at Saarland University.
We develop methods related to molecular dynamics simulations, with the aim to understand the relationship between structure, dynamics, and function of biological macromolecules.
We are hiring: a PhD position is available in computational membrane biophysics. Please apply until Feb. 9, 2026. Find out more.
We have several interesting Bachelor and Master projects available. Find out more.
Research Topics
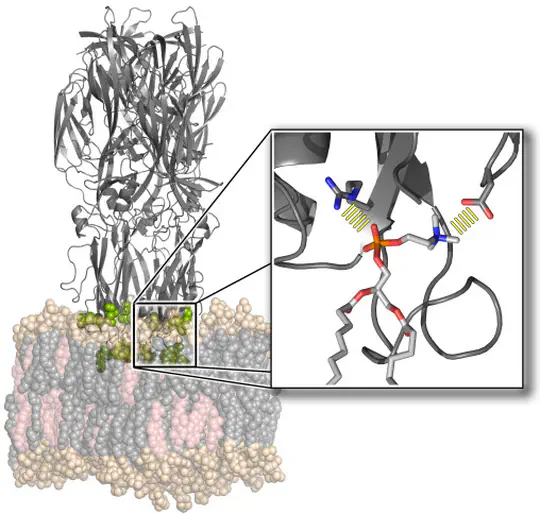
The function of biological membranes goes far beyond the formation of a mere barrier. Membranes are subject to ongoing structural remodeling, which is controlled by interactions with proteins and by the lipid composition. We develop free energy calculation techniques to understand how membrane composition and interactions with proteins (such as viral fusion proteins) enable functionally important events at membranes including membrane fusion, pore formation, or drug permeation.
Collecting experimental data is often difficult – but the interpretation of the data may be even more challenging, for instance because the information content of the experimental signals is low. We develop methods for combining MD simulations with experimental data to get the best of two worlds, with some focus on small-angle X-ray and neutron scattering data (SAXS/SANS). Our developments involve accurate SAXS/SANS predictions, protein structure and ensemble refinement, studies on the protein hydration shell, and modeling of experiments at X-ray free electron lasers. We share our methods via the web server WAXSiS and GROMACS-SWAXS.
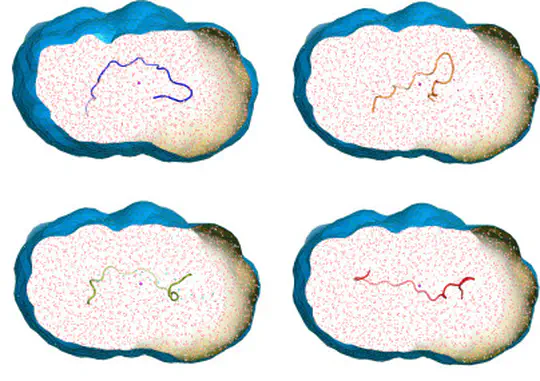
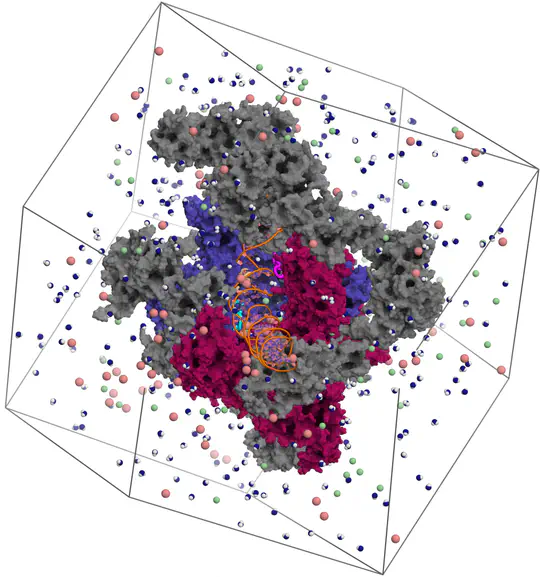
Proteins are not static building blocks but instead carry out their function –and malfunction– by structural transitions (Structure-function-dynamics relationship). We combine MD simulations with experiential data and enhanced-sampling techniques, to observe proteins while they function in atomic detail. Our portfolio comprises studies of molecular motors, protein-RNA/DNA complexes, membrane channels, and enzymes related to cancer progression.
Latest News
Latest Publications

We present BindFlow, a Python-based software for automated absolute binding free energy (ABFE) calculations at the free energy perturbation (FEP) or at the molecular mechanics Poisson–Boltzmann/generalized Born surface area [MM(PB/GB)SA] level of theory. BindFlow is free, open-source, user-friendly, and easily customizable, runs on workstations or distributed computing platforms, and provides extensive documentation and tutorials. BindFlow uses GROMACS as a molecular dynamics engine and provides built-in support for the small-molecule force fields GAFF, OpenFF, and Espaloma, as well as support for user-provided custom force fields. We test BindFlow by computing affinities for 139 receptor–ligand pairs, involving eight different targets, including six soluble proteins, one membrane protein, and one nonprotein host–guest system. We find that the agreement of BindFlow predictions with experiments is overall similar to gold standards in the field. Interestingly, we find that MM(PB/GB)SA achieves correlations that, for some systems and force fields, approach those obtained with FEP while requiring only a fraction of the computational cost. This study establishes BindFlow as a validated and accessible tool for ABFE calculations.
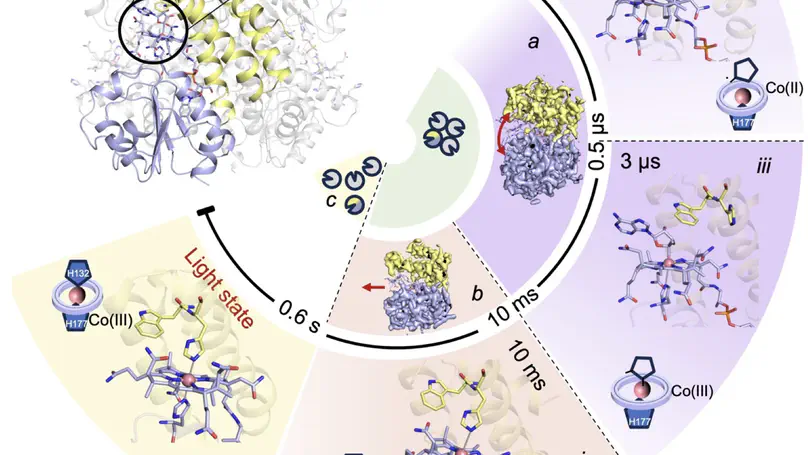
Photoreceptor proteins initiate, regulate and control fundamental biological processes such as vision, photosynthesis and circadian rhythms. A large photoreceptor subfamily uses vitamin B12 derivatives for light sensing, contrasting with the well-established mode of action of these organometallic derivatives in thermally activated enzymatic reactions. The molecular mechanism of B12 photoreception and how this differs to the thermal pathways remain unknown. Here we provide a detailed spatio-temporal description of photoactivation in the prototypical tetrameric B12 photoreceptor CarH from nanoseconds to seconds by using an integrative approach, combining time- and temperature-resolved structural and spectroscopic methods with quantum chemical calculations. High resolution structural snapshots of key intermediates illustrate how photocleavage of a Co–C bond triggers a pathway of structural changes that propagate throughout CarH from the B12 chromophore, via a previously unknown adduct, to finally cause tetramer dissociation. These unique intermediates, which differentiate CarH from thermally-activated B12 enzymes, steer the photoactivation pathway and act as the molecular bridge between photochemical and photobiological timescales. Our results offer a spatio-temporal understanding of CarH photoactivation and pave the way for designing and optimising B12-dependent photoreceptors for future optogenetic applications.

The hydration shell is an integral part of proteins since it plays key roles in conformational transitions, molecular recognition, and enzymatic activity. While the dynamics of the hydration shell have been described by spectroscopic techniques, the structure of the hydration shell remains less understood due to the lack of hydration shell-sensitive structural probes with high spatial resolution. We combined temperature-ramp small-angle X-ray scattering (T-ramp SAXS) from 255 to 335 K with molecular simulations to demonstrate that the hydration shells of the IgG-binding domain of Protein G (GB3) and the villin headpiece are remarkably temperature-sensitive. For proteins in the folded state, T-ramp SAXS data and explicit-solvent SAXS predictions consistently demonstrate decays of protein contrasts and radii of gyration with increasing temperature, which are shown to reflect predominantly temperature-sensitive, depleting hydration shells. The depletion is caused not merely by enhanced disorder within the hydration shells but also by partial displacements of surface-coordinated water molecules. Together, T-ramp SAXS and explicit-solvent SAXS calculations provide a novel structural view of the protein hydration shell, which underlies temperature-dependent processes such as cold denaturation, thermophoresis, or biomolecular phase separation.
Funding
Present and former