Computational Biophysics Group
Welcome to the Computational Biophysics Group at Saarland University.
We develop methods related to molecular dynamics simulations, with the aim to understand the relationship between structure, dynamics, and function of biological macromolecules.
We have several interesting Bachelor and Master projects available. Find out more.
Research Topics
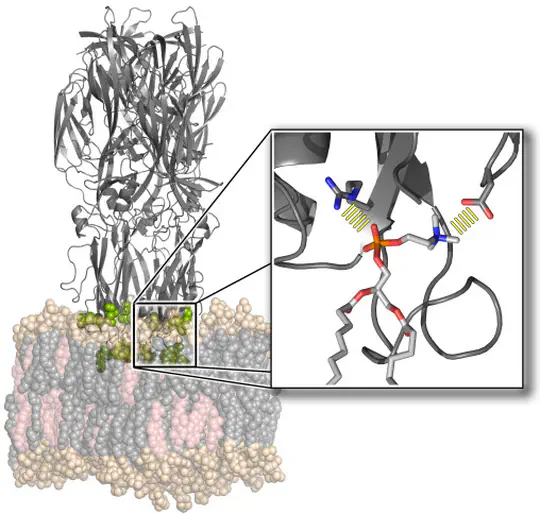
The function of biological membranes goes far beyond the formation of a mere barrier. Membranes are subject to ongoing structural remodeling, which is controlled by interactions with proteins and by the lipid composition. We develop free energy calculation techniques to understand how membrane composition and interactions with proteins (such as viral fusion proteins) enable functionally important events at membranes including membrane fusion, pore formation, or drug permeation.
Collecting experimental data is often difficult – but the interpretation of the data may be even more challenging, for instance because the information content of the experimental signals is low. We develop methods for combining MD simulations with experimental data to get the best of two worlds, with some focus on small-angle X-ray and neutron scattering data (SAXS/SANS). Our developments involve accurate SAXS/SANS predictions, protein structure and ensemble refinement, studies on the protein hydration shell, and modeling of experiments at X-ray free electron lasers. We share our methods via the web server WAXSiS and GROMACS-SWAXS.
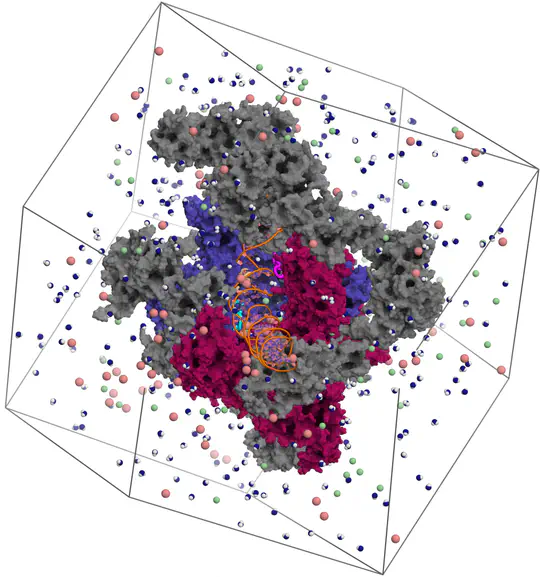
Proteins are not static building blocks but instead carry out their function –and malfunction– by structural transitions (Structure-function-dynamics relationship). We combine MD simulations with experiential data and enhanced-sampling techniques, to observe proteins while they function in atomic detail. Our portfolio comprises studies of molecular motors, protein-RNA/DNA complexes, membrane channels, and enzymes related to cancer progression.
Latest News
Latest Publications
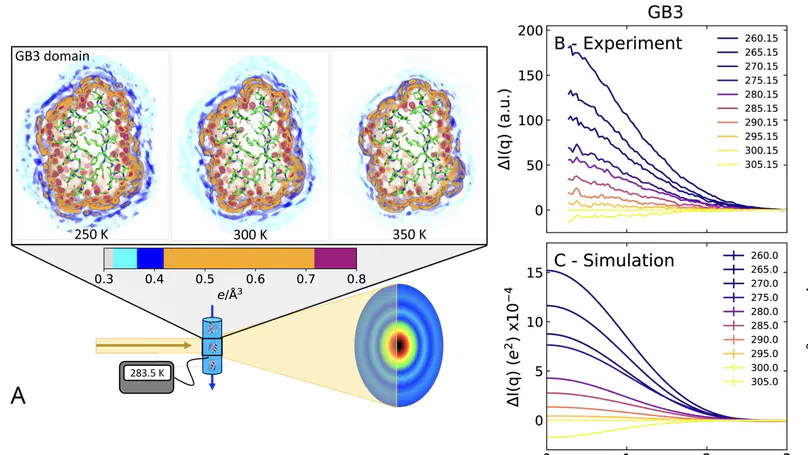
The hydration shell is an integral part of proteins since it plays key roles for conformational transitions, molecular recognition, and enzymatic activity. While the dynamics of the hydration shell have been described by spectroscopic techniques, the structure of the hydration shell remain less understood due to the lack of hydration shell-sensitive structural probes with high spatial resolution. We combined temperature-ramp small-angle X-ray scattering (T-ramp SAXS) from 255–335 K with molecular simulations to show that the hydration shells of the GB3 domain and villin headpiece are remarkably temperature-sensitive. For proteins in the folded state, T-ramp SAXS data and explicit-solvent SAXS predictions consistently demonstrate decays of protein contrasts and radii of gyration with increasing temperature, which are shown to reflect predominantly temperature-sensitive depleting hydration shells. The depletion is not merely caused by enhanced disorder within the hydration shells but also by partial displacements of surface-coordinated water molecules. Together, T-ramp SAXS and explicit-solvent SAXS calculations provide a novel structural view on the protein hydration shell, which underlies temperature-dependent processes such as cold denaturation, thermophoresis, or biomolecular phase separation.
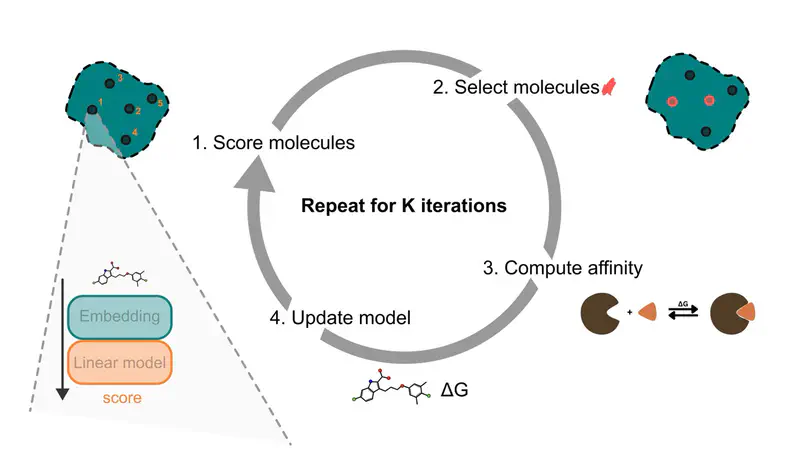
Predicting protein-ligand binding affinity with high accuracy is critical in structure-based drug discovery. While docking methods offer computational efficiency, they often lack the precision required for reliable affinity ranking. In contrast, molecular dynamics (MD)-based approaches such as MMGBSA provide more accurate binding free energy estimates but are computationally intensive, limiting their scalability. To address this trade-off, we introduce an active learning framework that automates molecule selection for docking and MD simulations, replacing manual expert-driven decisions with a data-efficient, model-guided strategy. Our approach integrates fixed - partly pre-trained deep learning - molecular embeddings (MolFormer, ChemBERTa-2, and Morgan fingerprints) with adaptive regression models (e.g. Bayesian Ridge and Random Forest) to iteratively improve binding affinity predictions. We evaluate this approach retrospectively on a new dataset of 60,000 chemically diverse compounds from ZINC-22 targeting the MCL1 protein using both AutoDock Vina and MMGBSA. Our results show that incorporating MMGBSA scores into the active learning loop significantly enhances performance, recovering 79.9% of the top 1% binders in the whole dataset, compared to only 6.7% when using docking scores alone. Notably, MMGBSA exhibits a stronger correlation with experimental binding affinities than AutoDock Vina on our dataset and enables more accurate ranking of candidate compounds in a runtime efficient way. Furthermore, we demonstrate that a one-at-a-time acquisition active learning strategy consistently outperforms traditional batched acquisition, the latter achieving just 78.4% recovery with MolFormer and Bayesian Ridge. These findings underscore the potential of integrating deep learning-based molecular representations with MD-level accuracy in an active learning framework, offering a scalable and efficient path to accelerate virtual screening and improve hit identification in drug discovery.
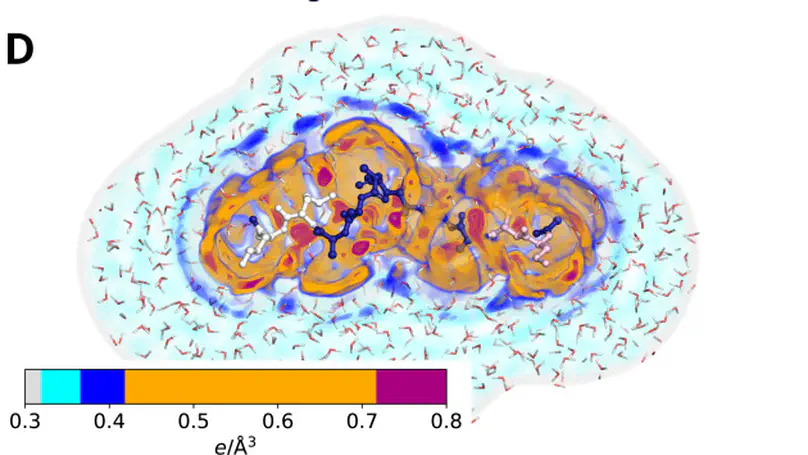
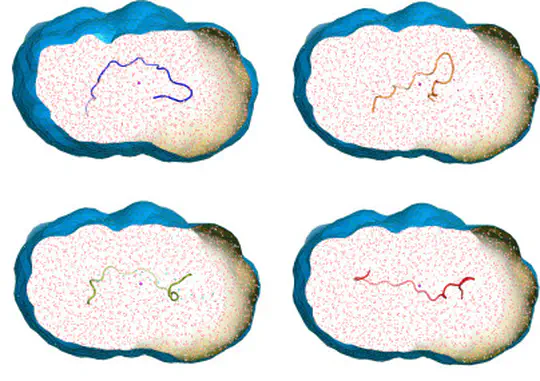
The protein hydration shell is a key mediator of processes such as molecular recognition, protein folding, and proton transfer. How surface-exposed amino acids shape the hydration shell structure is not well understood. We combine molecular dynamics simulations with explicit-solvent predictions of small-angle X-ray scattering (SAXS) curves to quantify the contributions of all 20 proteinogenic amino acids to the hydration shell of the globular GB3 domain and the intrinsically disordered protein (IDP) XAO. We focus on two quantities encoded by SAXS curves: the hydration shell effect on the radius of gyration and the electron density contrast between protein and solvent. We derive an amino-acid-specific contrast score, revealing that acidic residues generate the strongest contrast with 1 to 1.5 excess water molecules relative to alanine, followed by cationic and polar residues. In contrast, apolar residues generate a water depletion layer. These trends are consistent across simulations with different water models. Around the XAO peptide, the hydration shell is generally far weaker compared to the globular GB3 domain, indicating unfavorable water–peptide packing at the IDP surface. The hydration shell effect on the radius of gyration of the IDP is strongly conformation-dependent. Together, the calculations show that the composition and spatial arrangement of surface-exposed amino acids govern the hydration shell structure, with implications for a wide range of biological functions and for hydration-sensitive experimental techniques such as solution scattering.
Funding
Present and former